|
Genome editing using
the CRISPR/Cas-9 nucleases.
Genome editing has
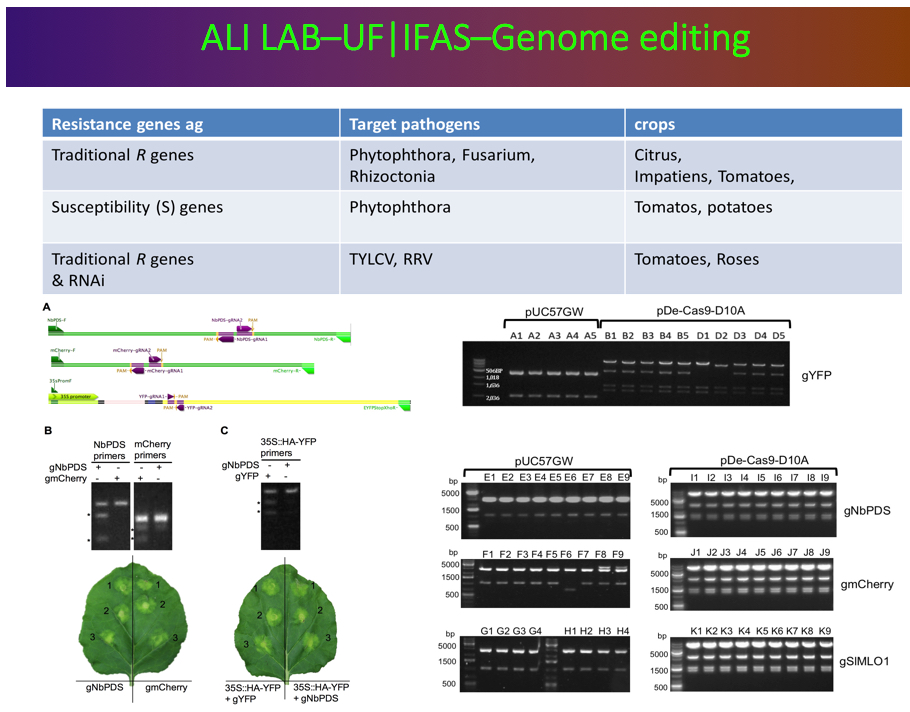
revolutionized almost all fields of biology. In our lab, we have developed
a robust genome editing method, which we are currently utilizing for
knocking out and knocking in several disease resistance genes in
economically important plants. In the future, we will be extending
this technology to fungi and oomycetes.
Plants use two major immunity
systems to defend themselves against pathogens. In one system,
conserved pathogen-associated molecular patterns (PAMP) are
recognized by plants through “Pattern Recognition
Receptors” (PRRs) leading to the activation of defense
responses. Pathogens inactivate PAMP-triggered immunity by delivering
virulent disease causing effectors proteins into cells. Plants
counteract by recognizing these effectors through resistance (R)
proteins, which leads to a more rapid and robust defense system
called effector-triggered immunity. Plant defenses include cell wall
reinforcement, hypersensitive response, and expression of many
defense genes including pathogenesis related (PR) genes. Introgression of
resistant genes from wild relatives using traditional breeding
methods is not efficient, especially in woody ornamentals, because of
difficulty in sexual hybridization and potential loss of desirable
traits. In addition, the gene-for-gene type R genes are in most cases rapidly
overcome by new virulent pathogen strains. Current crop transformations
for fungal resistance have been using individual defense genes, which
in most cases have resulted in partial resistance Durable and
broad-spectrum resistance can be accomplished with heterologously expressing multiple Pattern
Recognition Receptors (PRRs) along with other genes in one line. We
are transforming several PRRs
and other defense-related genes in unison into several economically
important crops including tomatoes, Geranium and roses. Since, our
approach is expected to provide broad-spectrum resistance, wherever
feasible, transgenic lines will be tested against other pathogens
too.
|